Section: New Results
RNA design
We extended our previous results on RNA design [29], obtained in collaboration with J. Hales, J. Manuch and L. Stacho (Simon Fraser University/Univ. British Columbia, Canada).
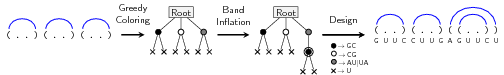
Our results provided complete characterizations for the structures that can be designed using restricted alphabets. We provided a complete characterization of designable structures without unpaired bases. When unpaired bases are allowed, we provided partial characterizations for classes of designable/undesignable structures, and showed that the class of designable structures is closed under the stutter operation. Membership of a given structure to any of the classes can be tested in linear time and, for positive instances, a solution could be found in linear time. Finally, we considered a structure-approximating version of the problem that allows to extend helices and, assuming that the input structure avoids two motifs, we provided a linear-time algorithm that produces a designable structure with at most twice more base pairs than the input structure, as illustrated by Fig. 3.
|
In a manuscript accepted for publication in Algorithmica [4], we have shown that our previous results [29] hold for more sophisticated energy models where base-pairs are associated with arbitrary energy contributions. This result, which required a complete overhaul of our previous proofs (e.g. using arguments based on graph coloring), allows us to foresee an extension of (at least some of) our results to state-of-the-art models, such as the Turner energy model.
We also initiated a collaboration with Danny Barash's group at Ben-Gurion university (Israel). We contributed a review of existing tools and techniques for RNA design, to appear as an article within the Briefings in Bioinformatics series [2]. We also combined previously contributed methods for design into a new method and web-server for the design of RNAs [3]. This collaboration stemed from the observation that IncaRNAtion [36], a random generation algorithm for RNA design recently developed in collaboration with Jérôme Waldispühl's group at McGill University (Montreal, Canada), produced excellent starting points (seed) for classic algorithms based on local-search. In particular, the combination of IncaRNAtion and RNAfbInv [45] was found to yield particularly promising candidates for design.